No feedback found for this session
SQL joins
SQL
beginner
NoteAbout this course
- This course is designed as a basic introduction to SQL delivered as live interactive sessions on Teams
- It’s intended as a light introduction for users with good general digital skills (broadly level 2 in the Digital and Data Capability Framework)
- It’s heavily based on the excellent W3 schools introductory SQL course
- If you’re looking for a resource to use to support independent study, W3 schools is a better option than this page
- this material is largely intended to support our live interactive training sessions
- we use posit.cloud for this course. Although that platform is mainly meant for analysts writing R code, we can trick it to allow us to practice our SQL skills
About this session
This is a session about joins in SQL:
- we’ll introduce joins in general terms
- we’ll then go on to talk about different types of join in SQL
- then we’ll do lots of practice of basic joins
- and finish with a couple of helpful points for simplifying your work with joins
Setup
- You will need a free posit.cloud account. Please set this up and check that you have access before the session begins
- Once you’ve logged-in to posit.cloud, please create a new Rstudio project from Github Repository
- Supply the following URL when prompted: https://github.com/NES-DEW/KIND_sql_intro/
- Once the project is deployed, clear your Environment (broom icon in the environment pane)
- Then open the
s4_setup.Rscript by from the bottom-right hand files pane - Click the
Sourcebutton at the top right-hand corner of the source pane
- Then open the
s4_joins.sqlscript from the files pane, andCtrl+Shift+Entertwice to preview your data for this session
NoteManual setup instructions
- You will need a free posit.cloud account. Please set this up and check that you have access before the session begins
- Once you’ve logged-in to posit.cloud, please create a new Rstudio project
- In that project, create a new R script:
Data
- This session uses some sample data, which we’ll load directly from the web in our R script
- Copy and paste the following R code into that script:
pkg <- c("dbplyr", "RSQLite", "dplyr", "readr")
install.packages(setdiff(pkg, rownames(installed.packages())))
library(dbplyr)
library(dplyr)
library(readr)
read_rds("https://nes-dew.github.io/KIND-training/skills/data/prescribing_data.rds") |>
list2env(.GlobalEnv)
conn <- src_memdb() # create an sqlite db in memory
copy_to(conn, ab_city_presc, overwrite = T)
copy_to(conn, bnf_data, overwrite = T)
copy_to(conn, gp_practices, overwrite = T)
copy_to(conn, hbt, overwrite = T)
copy_to(conn, practice_names, overwrite = T)
copy_to(conn, practice_lists, overwrite = T)Creating our SQL database
- save your script and click the
Sourcebutton- that will create a simple SQL database in memory
- now create a new SQL script (again, from the
Filemenu) - delete all the pre-populated lines of code, as follows:
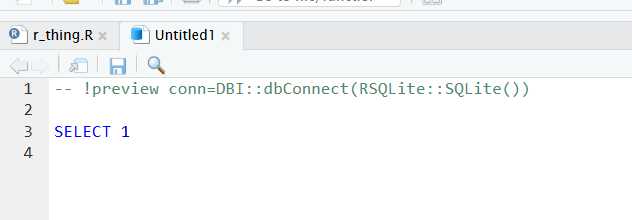
- to connect with our SQL db in memory, add the following line to the head of your SQL script:
-- !preview conn=src_memdb()$conData structure
- we’ve now got a series of SQL tables in memory
- in one way and another those tables are all about pharmaceutical prescribing
- we’ll start with two very simple tables, before moving on to experiment with some more real-world data
- the first,
practice_namescontains the names and ID codes for a few GP practices:
SELECT * FROM practice_names;| PracticeCode | GPPracticeName |
|---|---|
| 30951 | Rubislaw Place Medical Group |
| 30909 | Albyn Medical Practice |
| 31206 | Scotstown Medical Group |
| 30627 | Links Medical Practice |
- the second contains some practice list sizes for these GP practices - which tell you how many patients are registered for each practice
SELECT * FROM practice_lists;| PracticeCode | PracticeListSize |
|---|---|
| 30951 | 8038 |
| 30909 | 11891 |
| 31206 | 12344 |
A first join
- for example, we could take our
practice_namestable, and join on the additional column found in thepractice_liststable:
SELECT * FROM practice_names
JOIN practice_lists
ON practice_names.PracticeCode = practice_lists.PracticeCode
-- take our practice_names table, and for every matching PracticeCode bring in the other column(s) from practice_lists| PracticeCode | GPPracticeName | PracticeCode | PracticeListSize |
|---|---|---|---|
| 30951 | Rubislaw Place Medical Group | 30951 | 8038 |
| 30909 | Albyn Medical Practice | 30909 | 11891 |
| 31206 | Scotstown Medical Group | 31206 | 12344 |
- to note:
- there are two parts to a join. A join keyword, like
LEFT JOINwhich determines the logic of the join -
ONwhich describes how the columns in our data should relate to one another. Note thetable_name.column_namesyntax. Call that the key
- there are two parts to a join. A join keyword, like
- we can do several kinds of SQL join by using a different join keyword
- for a table
xto which we’re joiningy:-
JOIN: includes all rows in x and y. Also known asINNER JOIN
-
Types of join
An inner join is not the only way we can join in SQL. We can also:
-
LEFT JOIN: includes all rows in x
-
RIGHT JOIN: includes all rows in y
-
FULL JOIN: includes all rows in x or y
If a row in x matches multiple rows in y, all the rows in y will be returned once for each matching row in x.
Prescribing data
- as well as our practice_names and practice_lists tables, we’ve got some more interesting data set up in our SQL database about prescriptions
NoteExercise
- inspect the following tables:
gp_practiceshbtab_city_prescbnf_data
- there are several possible joins we might attempt here: please make a suggestion about a join you’d be interesting in trying in the chat
A starting join
- look at the
ab_city_presctable, which is contains data about prescriptions in the Aberdeen City area - this is hard to read because the drug information in the
BNFItemCodecolumn is expressed as codes (like1202010Y0BBAAAA) rather than drug names - luckily,
bnf_datais a lookup: the corresponding column in that table isbnf_presentation_code
SELECT * FROM ab_city_presc JOIN bnf_data ON ab_city_presc.BNFItemCode = bnf_data.bnf_presentation_code;| GPPractice | PaidDateMonth | BNFItemCode | PrescribedType | NumberOfPaidItems | PaidQuantity | GrossIngredientCost | bnf_presentation_code | bnf_chapter | bnf_chapter_code | bnf_section | bnf_section_code | bnf_paragraph | bnf_paragraph_code | bnf_subparagraph | bnf_subparagraph_code | bnf_chemical_substance | bnf_chemical_substance_code | bnf_product | bnf_product_code | bnf_presentation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 30030 | 19875 | 1202010U0AAAAAA | VMP | 39 | 6160 | 404.80 | 1202010U0AAAAAA | Ear, Nose and Oropharynx | 12 | Drugs acting on the nose | 1202 | Drugs used in nasal allergy | 120201 | Drugs used in nasal allergy | 1202010 | Mometasone furoate | 1202010U0 | Mometasone furoate (Nasal) | 1202010U0AA | Mometasone 50micrograms/dose nasal spray |
| 30030 | 19875 | 1203010E0AAABAB | VMP | 3 | 90 | 9.63 | 1203010E0AAABAB | Ear, Nose and Oropharynx | 12 | Drugs acting on the oropharynx | 1203 | Drugs for oral ulceration and inflammation | 120301 | Drugs for oral ulceration and inflammation | 1203010 | Benzydamine hydrochloride | 1203010E0 | Benzydamine hydrochloride (Mouthwash) | 1203010E0AA | Benzydamine 0.15% oromucosal spray sugar free |
| 30030 | 19875 | 1202010W0BBAAAA | AMP | 3 | 360 | 19.32 | 1202010W0BBAAAA | Ear, Nose and Oropharynx | 12 | Drugs acting on the nose | 1202 | Drugs used in nasal allergy | 120201 | Drugs used in nasal allergy | 1202010 | Fluticasone furoate | 1202010W0 | Avamys | 1202010W0BB | Avamys 27.5micrograms/dose nasal spray |
| 30030 | 19875 | 1202010W0BBAAAA | VMP | 7 | 1200 | 64.40 | 1202010W0BBAAAA | Ear, Nose and Oropharynx | 12 | Drugs acting on the nose | 1202 | Drugs used in nasal allergy | 120201 | Drugs used in nasal allergy | 1202010 | Fluticasone furoate | 1202010W0 | Avamys | 1202010W0BB | Avamys 27.5micrograms/dose nasal spray |
| 30030 | 19875 | 1203010E0BBAAAA | AMP | 1 | 300 | 6.50 | 1203010E0BBAAAA | Ear, Nose and Oropharynx | 12 | Drugs acting on the oropharynx | 1203 | Drugs for oral ulceration and inflammation | 120301 | Drugs for oral ulceration and inflammation | 1203010 | Benzydamine hydrochloride | 1203010E0 | Difflam (Mouthwash) | 1203010E0BB | Difflam Oral Rinse 0.15% solution |
| 30030 | 19875 | 1202010Y0BBAAAA | AMP | 8 | 1200 | 148.00 | 1202010Y0BBAAAA | Ear, Nose and Oropharynx | 12 | Drugs acting on the nose | 1202 | Drugs used in nasal allergy | 120201 | Drugs used in nasal allergy | 1202010 | Fluticasone propionate/azelastine hydrochloride (Nasal) | 1202010Y0 | Dymista | 1202010Y0BB | Dymista 137micrograms/dose / 50micrograms/dose nasal spray |
| 30030 | 19875 | 1202010Y0BBAAAA | VMP | 1 | 120 | 14.80 | 1202010Y0BBAAAA | Ear, Nose and Oropharynx | 12 | Drugs acting on the nose | 1202 | Drugs used in nasal allergy | 120201 | Drugs used in nasal allergy | 1202010 | Fluticasone propionate/azelastine hydrochloride (Nasal) | 1202010Y0 | Dymista | 1202010Y0BB | Dymista 137micrograms/dose / 50micrograms/dose nasal spray |
| 30030 | 19875 | 1201010Q0BCAAAC | AMP | 1 | 10 | 7.45 | 1201010Q0BCAAAC | Ear, Nose and Oropharynx | 12 | Drugs acting on the ear | 1201 | Otitis externa | 120101 | Otitis externa | 1201010 | Hydrocortisone | 1201010Q0 | Otosporin | 1201010Q0BC | Otosporin ear drops |
| 30030 | 19875 | 1202020L0AACCCC | VMP | 1 | 20 | 9.50 | 1202020L0AACCCC | Ear, Nose and Oropharynx | 12 | Drugs acting on the nose | 1202 | Topical nasal decongestants | 120202 | Topical nasal decongestants | 1202020 | Sodium chloride | 1202020L0 | Sodium chloride (Nasal) | 1202020L0AA | Sodium chloride 0.9% neb liq 2.5ml unit dose ampoules |
| 30030 | 19875 | 120101050BCAAAB | AMP | 4 | 20 | 13.08 | 120101050BCAAAB | Ear, Nose and Oropharynx | 12 | Drugs acting on the ear | 1201 | Otitis externa | 120101 | Otitis externa | 1201010 | Dexamethasone | 120101050 | Otomize | 120101050BC | Otomize ear spray |
Reminder!
- you can subset the columns returned by a query via
SELECT:
SELECT GPPractice, PaidDateMonth, PaidQuantity, bnf_presentation
FROM ab_city_presc
JOIN bnf_data
ON ab_city_presc.BNFItemCode = bnf_data.bnf_presentation_code;| GPPractice | PaidDateMonth | PaidQuantity | bnf_presentation |
|---|---|---|---|
| 30030 | 19875 | 6160 | Mometasone 50micrograms/dose nasal spray |
| 30030 | 19875 | 90 | Benzydamine 0.15% oromucosal spray sugar free |
| 30030 | 19875 | 360 | Avamys 27.5micrograms/dose nasal spray |
| 30030 | 19875 | 1200 | Avamys 27.5micrograms/dose nasal spray |
| 30030 | 19875 | 300 | Difflam Oral Rinse 0.15% solution |
| 30030 | 19875 | 1200 | Dymista 137micrograms/dose / 50micrograms/dose nasal spray |
| 30030 | 19875 | 120 | Dymista 137micrograms/dose / 50micrograms/dose nasal spray |
| 30030 | 19875 | 10 | Otosporin ear drops |
| 30030 | 19875 | 20 | Sodium chloride 0.9% neb liq 2.5ml unit dose ampoules |
| 30030 | 19875 | 20 | Otomize ear spray |
The AS keyword
- as you might have noticed above, you end up writing the names of tables a couple of times during a join
- our tables have slightly long and tricky names
- you can temporarily rename tables using
AS:
SELECT GPPractice, PaidDateMonth, PaidQuantity, bnf_presentation
FROM ab_city_presc AS ab
JOIN bnf_data AS bnf
ON ab.BNFItemCode = bnf.bnf_presentation_code;| GPPractice | PaidDateMonth | PaidQuantity | bnf_presentation |
|---|---|---|---|
| 30030 | 19875 | 6160 | Mometasone 50micrograms/dose nasal spray |
| 30030 | 19875 | 90 | Benzydamine 0.15% oromucosal spray sugar free |
| 30030 | 19875 | 360 | Avamys 27.5micrograms/dose nasal spray |
| 30030 | 19875 | 1200 | Avamys 27.5micrograms/dose nasal spray |
| 30030 | 19875 | 300 | Difflam Oral Rinse 0.15% solution |
| 30030 | 19875 | 1200 | Dymista 137micrograms/dose / 50micrograms/dose nasal spray |
| 30030 | 19875 | 120 | Dymista 137micrograms/dose / 50micrograms/dose nasal spray |
| 30030 | 19875 | 10 | Otosporin ear drops |
| 30030 | 19875 | 20 | Sodium chloride 0.9% neb liq 2.5ml unit dose ampoules |
| 30030 | 19875 | 20 | Otomize ear spray |
Multiple joins
- say you’d like more information about the GP practices. That can be brought in from the
gp_practicestable with a second join:
SELECT GPPracticeName, PaidDateMonth, PaidQuantity, bnf_presentation
FROM ab_city_presc AS ab
JOIN bnf_data AS bnf
ON ab.BNFItemCode = bnf.bnf_presentation_code
JOIN gp_practices AS gp ON ab.GPPractice = gp.PracticeCode| GPPracticeName | PaidDateMonth | PaidQuantity | bnf_presentation |
|---|---|---|---|
| Garthdee Medical Group | 20148 | 8 | Sofradex ear/eye drops |
| Garthdee Medical Group | 20179 | 20 | Sofradex ear/eye drops |
| Garthdee Medical Group | 20209 | 10 | Sofradex ear/eye drops |
| Garthdee Medical Group | 19875 | 20 | Otomize ear spray |
| Garthdee Medical Group | 19905 | 5 | Otomize ear spray |
| Garthdee Medical Group | 19936 | 15 | Otomize ear spray |
| Garthdee Medical Group | 19967 | 5 | Otomize ear spray |
| Garthdee Medical Group | 19997 | 10 | Otomize ear spray |
| Garthdee Medical Group | 20028 | 30 | Otomize ear spray |
| Garthdee Medical Group | 20058 | 5 | Otomize ear spray |
Prescribing data info
Show data creation script
# this script takes open data about prescribing in Scotland, and produces a small and interesting subset of ENT prescriptions from Aberdeen City GPs June 2024-May 2025
# it's designed purely for training purposes. In the KIND network we're using it for our introductory SQL course, for example.
# this dataset starts with the twelve monthly files by prescriber location covering June 2024-May 2025: https://www.opendata.nhs.scot/dataset/prescriptions-in-the-community. These are downloaded locally - see note below
# you can find supporting information in the data glossary https://publichealthscotland.scot/media/34209/open_data_glossary_of_terms.pdf
# you may also find the openprescribing site helpful about BNF codes: https://openprescribing.net/bnf/040702/
# and more BNF info from the NHSBSA website - https://applications.nhsbsa.nhs.uk/infosystems/welcome (guest login needed but free and open to all)
library(dplyr)
library(readr)
## bnf info ----
bnf <- "https://opendata.nhsbsa.net/dataset/29d25de3-02cd-4755-9dee-cdc37e37b5f3/resource/b1ce568d-37c4-4dd6-8bf4-57e4b044bf6c/download/bnf_code_current_202507_version_88.csv"
bnf_data <- read_csv(bnf) |>
filter(BNF_CHAPTER == "Ear, Nose and Oropharynx") |>
rename_with(tolower) |>
select(-year_month) |>
relocate(bnf_presentation_code)
ent <- bnf_data |>
pull(bnf_presentation_code) # subset of ENT-specific codes
## get the GP practice info for Aberdeen city----
jul_2025 <- "https://www.opendata.nhs.scot/dataset/f23655c3-6e23-4103-a511-a80d998adb90/resource/30b06220-17ad-44e8-b6c5-658d41ec1ea5/download/practice_contactdetails_jul2025-open-data.csv"
gp_practices <- read_csv(jul_2025) |>
filter(str_detect(GPCluster, "Aberdeen City"))
gps <- gp_practices |>
pull(PracticeCode)
## get the health board lookup----
hbt <- read_csv("https://www.opendata.nhs.scot/dataset/9f942fdb-e59e-44f5-b534-d6e17229cc7b/resource/652ff726-e676-4a20-abda-435b98dd7bdc/download/hb14_hb19.csv") |>
select(HB, HBName)
## build the dataset ----
ab_city_presc <- read_csv(list.files("data/community_presc/", full.names = T)) |> # directory of monthly files downloaded from https://www.opendata.nhs.scot/dataset/prescriptions-in-the-community
filter(GPPractice %in% gps) |>
filter(BNFItemCode %in% ent) |>
select(GPPractice, PaidDateMonth, BNFItemCode, PrescribedType, NumberOfPaidItems, PaidQuantity, GrossIngredientCost) |>
mutate(PaidDateMonth = ym(PaidDateMonth)) |>
mutate(PaidDateMonth = as.character(PaidDateMonth))
ab_mini <- gp_practices |>
select(PracticeCode, GPPracticeName, PracticeListSize) |>
slice_sample(n = 3)
practice_names <- ab_mini |>
select(-PracticeListSize) |>
bind_rows(tibble(PracticeCode = 30627,
GPPracticeName = "Links Medical Practice"))
practice_lists <- ab_mini |>
select(-GPPracticeName)
practice_names |>
inner_join(practice_lists)
list(ab_city_presc = ab_city_presc, bnf_data = bnf_data, gp_practices = gp_practices, hbt = hbt, practice_lists = practice_lists, practice_names = practice_names) |>
write_rds("data/prescribing_data.rds")SQL query summary
Code
SELECT * FROM practice_names; -- to preview practice names
SELECT * FROM practice_lists; -- and list sizes
SELECT * FROM practice_names
JOIN practice_lists
ON practice_names.PracticeCode = practice_lists.PracticeCode -- to join list size to practice info
SELECT * FROM ab_city_presc JOIN bnf_data ON ab_city_presc.BNFItemCode = bnf_data.bnf_presentation_code; -- to join drug information into Aberdeen City prescribing data
SELECT GPPractice, PaidDateMonth, PaidQuantity, bnf_presentation
FROM ab_city_presc
JOIN bnf_data
ON ab_city_presc.BNFItemCode = bnf_data.bnf_presentation_code; -- join then subset
SELECT GPPractice, PaidDateMonth, PaidQuantity, bnf_presentation
FROM ab_city_presc AS ab
JOIN bnf_data AS bnf
ON ab.BNFItemCode = bnf.bnf_presentation_code; -- join with temporary renaming via AS
SELECT GPPracticeName, PaidDateMonth, PaidQuantity, bnf_presentation
FROM ab_city_presc AS ab
JOIN bnf_data AS bnf
ON ab.BNFItemCode = bnf.bnf_presentation_code
JOIN gp_practices AS gp ON ab.GPPractice = gp.PracticeCode -- multiple joins Acknowledgements
Like all our courses and sessions, this is a team effort, and I thank the network as a whole for their encouragements and contributions. Specific thanks in this case go to Ben Harley (PHS), Amanda King (NHS GGC), Steven Knapman (NHS Fife), and James McMahon (Public Health Scotland), and to the members of the pilot cohort for this session.