Summarising data with dplyr
Previous attendees have said…
- 7 previous attendees have left feedback
- 86% would recommend this session to a colleague
- 100% said that this session was pitched correctly
- Great session, providing practical use cases of key concepts around dplyr summarise (and why you should use reframe instead), mutate, group by and so much more. This session provided me with clear understanding of the differences between these functions and when/how to use them. As always, highly recommended session.
- Pitched perfectly to my level, not easy to find training at this level, really useful, extremely relevant to my work as well as highly enjoyable, so yes, cannot recommend the course highly enough, Brendan is a rockstar trainer.
- I ended up pretty confused in this session. I use count a lot and summarise never use it so was wanting tidy up my understanding, however the diversions to different code confused me as I’m a slow typer. I do now get group_by etc and I liked the add ons .
Session outline
This session is an 🌶🌶 intermediate practical designed for those with some R experience. The aim of this session is to do three things with dplyr:
- show how to approach summarising data
- explain how grouping works
- show some simple summary functions
You might also like some of the other dplyr-themed practical sessions:
Setup
Let’s do a little bit of package loading, find some interesting data, and then start summarising things:
Data loading
For the first half of the session, we’ll use a real dataset that shows deaths by socioeconomic deprivation. Full details on the PHS opendata page:
SMR_SIMD <- read_csv("https://www.opendata.nhs.scot/datastore/dump/e6849f09-3a5c-44c6-8029-260882345071?bom=True") |>
select(-c(`_id`, contains("QF"), Country)) |>
tidyr::separate_wider_delim(TimePeriod, "Q", names = c("Year", "Quarter")) |>
mutate(Year = as.numeric(Year),
Quarter = as.numeric(Quarter) * 3) |>
mutate(Date = ymd(paste(Year, Quarter, "01"))) |>
mutate(SIMDQuintile = case_when(SIMDQuintile == "1 - most deprived" ~ "1",
SIMDQuintile == "5 - least deprived" ~ "5",
TRUE ~ SIMDQuintile)) |>
relocate(last_col()) Data preview
We’ll do a quick skim() of the data to help you get a better sense of it:
skimr::skim(SMR_SIMD)| Name | SMR_SIMD |
| Number of rows | 222 |
| Number of columns | 7 |
| _______________________ | |
| Column type frequency: | |
| character | 1 |
| Date | 1 |
| numeric | 5 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| SIMDQuintile | 37 | 0.83 | 1 | 1 | 0 | 5 | 0 |
Variable type: Date
| skim_variable | n_missing | complete_rate | min | max | median | n_unique |
|---|---|---|---|---|---|---|
| Date | 0 | 1 | 2016-09-01 | 2025-09-01 | 2021-03-01 | 37 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| Year | 0 | 1 | 2020.62 | 2.70 | 2016.00 | 2018.00 | 2021.00 | 2023.00 | 2025.00 | ▆▇▇▇▇ |
| Quarter | 0 | 1 | 7.54 | 3.32 | 3.00 | 6.00 | 9.00 | 9.00 | 12.00 | ▇▇▁▇▇ |
| NumberOfDeaths | 0 | 1 | 1139.43 | 559.83 | 7.00 | 1005.00 | 1285.00 | 1544.50 | 2056.00 | ▃▁▆▇▂ |
| NumberOfPatients | 0 | 1 | 35551.51 | 16873.66 | 490.00 | 33593.25 | 40099.50 | 46094.50 | 59076.00 | ▃▁▃▇▃ |
| CrudeRate | 0 | 1 | 3.00 | 0.79 | 1.04 | 2.72 | 3.03 | 3.37 | 5.58 | ▂▃▇▁▁ |
This dataset is especially good for practising summarising, because there are various different plausible groups that we might like to investigate in it - especially the intersection between SIMDQuintiles (indicating different levels of deprivation) with the various date-based year/season/month groups that might be of interest for health improvement work:
Example time-series
SMR_SIMD |>
ggplot() +
geom_line(aes(x = Date, y = NumberOfDeaths, colour = SIMDQuintile)) +
theme_minimal()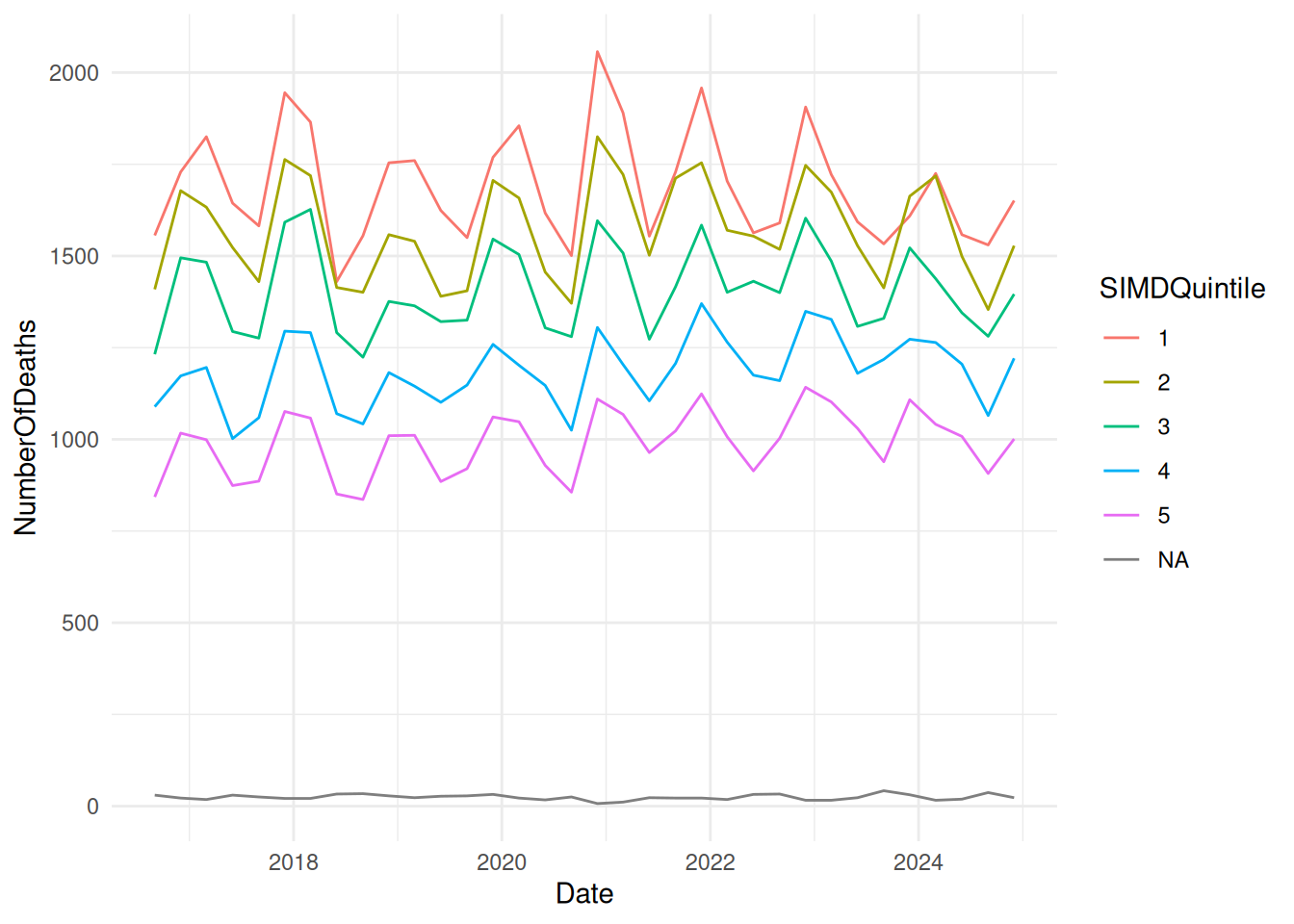
summarise()
This is the standard way of summarising data in dplyr. The description on the dplyr reference page for summarise is admirably clear:
summarise()creates a new data frame. It returns one row for each combination of grouping variables; if there are no grouping variables, the output will have a single row summarising all observations in the input. It will contain one column for each grouping variable and one column for each of the summary statistics that you have specified.
Basic example
group_by()
summarise() is especially strong in concert with group_by():
Care with groups
Two important things to note here:
-
group_bydoesn’t change how the data looks - just how it behaves: - Each call to
summarise()removes a layer of grouping
Renaming summary columns
You can build simple formulae inside summarise():
Expressions within summaries
You can also build-up more complex expressions within summarise
# A tibble: 10 × 2
Year surviving
<dbl> <dbl>
1 2016 447292
2 2017 888462
3 2018 885537
4 2019 919377
5 2020 662277
6 2021 737799
7 2022 778773
8 2023 832949
9 2024 851898
10 2025 635119by()
You can, in another recent change, group inside the summarise itself via .by():
# A tibble: 10 × 2
Year surviving
<dbl> <dbl>
1 2016 447292
2 2017 888462
3 2018 885537
4 2019 919377
5 2020 662277
6 2021 737799
7 2022 778773
8 2023 832949
9 2024 851898
10 2025 635119This always returns an ungrouped tibble - so important to know that it’s not a direct substitute for an ordinary group_by()…
summarise() removes one layer of grouping
The most confusing aspect of summarise() is that it removes the bottom layer of grouping each time. Here, we start with our data grouped by Year and Quarter. After summarising, the data is grouped by Year only.:
SMR_SIMD |>
group_by(Year, Quarter) |>
summarise(att = sum(NumberOfDeaths)) |>
group_vars()[1] "Year"Tools for understanding groupings
group_vars() is just one of a group of functions in dplyr for understanding grouping metadata. Let’s start with some simple grouped data. We can discover the groups that we’re working with using groups():
group_vars()
Simpler information is produced by group_vars():
SMR_SIMD |>
group_by(Year) |>
group_vars() # vector[1] "Year"
group_data() etc
Much fuller information by group_data(), group_rows() and friends:
SMR_SIMD |>
group_by(Year) |>
group_data() # tibble# A tibble: 10 × 2
Year .rows
<dbl> <list<int>>
1 2016 [12]
2 2017 [24]
3 2018 [24]
4 2019 [24]
5 2020 [24]
6 2021 [24]
7 2022 [24]
8 2023 [24]
9 2024 [24]
10 2025 [18]SMR_SIMD |>
group_by(Year) |>
group_rows() # list of each group's rows<list_of<integer>[10]>
[[1]]
[1] 1 2 3 4 5 6 7 8 9 10 11 12
[[2]]
[1] 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
[[3]]
[1] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60
[[4]]
[1] 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84
[[5]]
[1] 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103
[20] 104 105 106 107 108
[[6]]
[1] 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127
[20] 128 129 130 131 132
[[7]]
[1] 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151
[20] 152 153 154 155 156
[[8]]
[1] 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175
[20] 176 177 178 179 180
[[9]]
[1] 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199
[20] 200 201 202 203 204
[[10]]
[1] 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222reframe()
A recent change in dplyr 1.1.0 is that summarise() now will only return one row per group. A new function, reframe(), has been developed to produce multiple-row summaries. It works exactly like summarise() except, rather than removing one grouping layer per operation, it always returns an ungrouped tibble. The syntax is the same as summarise():
sum <- ae_attendances |>
group_by(year = lubridate::floor_date(period, unit = "year")) |>
summarise(
year = lubridate::year(year),
non_admissions = sum(attendances - admissions)
)
ref <- ae_attendances |>
group_by(year = lubridate::floor_date(period, unit = "year")) |>
reframe(
year = lubridate::year(year),
non_admissions = sum(attendances - admissions)
)
waldo::compare(sum, ref)`class(old)`: "grouped_df" "tbl_df" "tbl" "data.frame"
`class(new)`: "tbl_df" "tbl" "data.frame"
`attr(old, 'groups')` is an S3 object of class <tbl_df/tbl/data.frame>, a list
`attr(new, 'groups')` is absentcount()
count() counts unique values, and is equivalent to using group_by() and then summarise():
synthetic_news_data |>
count(died) # A tibble: 2 × 2
died n
<int> <int>
1 0 930
2 1 70
count() is a shorthand for grouping and summarising
That’s roughly equivalent to:
tally()
tally() works similarly, except without the group_by():
Weighted counts
A possible source of confusion is that adding a column name to either count() or tally() performs a weighted count of that column:
synthetic_news_data |>
tally(age) # totally dis-similar behaviour to count# A tibble: 1 × 1
n
<int>
1 69647Roughly equivalent to:
Named arguments
Personally, it seems wise to use named arguments inside count() and tally() to make sure it’s obvious what the column name is doing:
synthetic_news_data |>
count(wt = age)# A tibble: 1 × 1
n
<int>
1 69647Sorting
count() has a useful sort option to arrange by group size:
synthetic_news_data |>
count(syst, sort=T) # A tibble: 125 × 2
syst n
<int> <int>
1 130 28
2 120 26
3 139 26
4 150 25
5 135 24
6 126 22
7 134 22
8 119 21
9 123 21
10 112 20
# ℹ 115 more rowsControlling column names
And you can name your new count column something more descriptive than n:
synthetic_news_data |>
count(syst, name = "count_of_something") # A tibble: 125 × 2
syst count_of_something
<int> <int>
1 65 2
2 76 2
3 78 1
4 80 6
5 81 1
6 82 2
7 83 1
8 85 5
9 86 2
10 87 2
# ℹ 115 more rowsEmpty groups
If you want to count empty groups, use .drop:
# A tibble: 2 × 2
male n
<fct> <int>
1 0 524
2 1 476# A tibble: 3 × 2
male n
<fct> <int>
1 0 524
2 1 476
3 2 0add_count()
Both count() and tally() have add_ variants, which work like mutate() in that they add a new column containing the count:
add_tally()
add_tally() gives a col with the same value in each row of a group:
# A tibble: 6 × 13
# Groups: died [2]
male age NEWS syst dias temp pulse resp sat sup alert died n
<int> <int> <int> <int> <int> <dbl> <int> <int> <int> <int> <int> <int> <int>
1 0 68 3 150 98 36.8 78 26 96 0 0 0 930
2 1 94 1 145 67 35 62 18 96 0 0 0 930
3 0 85 0 169 69 36.2 54 18 96 0 0 0 930
4 0 80 5 130 81 37 129 18 97 0 0 1 70
5 0 62 6 108 65 34.4 43 18 99 0 0 1 70
6 1 64 0 114 82 36.2 78 18 97 0 0 1 70rowwise()
rowwise() forms groups per row. For example, we could take the average of the three columns in ae_attendances:
# A tibble: 12,765 × 7
# Rowwise:
period org_code type attendances breaches admissions mean
<date> <fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 2017-03-01 RF4 1 21289 2879 5060 9743.
2 2017-03-01 RF4 2 813 22 0 278.
3 2017-03-01 RF4 other 2850 6 0 952
4 2017-03-01 R1H 1 30210 5902 6943 14352.
5 2017-03-01 R1H 2 807 11 0 273.
6 2017-03-01 R1H other 11352 136 0 3829.
7 2017-03-01 AD913 other 4381 2 0 1461
8 2017-03-01 RYX other 19562 258 0 6607.
9 2017-03-01 RQM 1 17414 2030 3597 7680.
10 2017-03-01 RQM other 7817 86 0 2634.
# ℹ 12,755 more rowsmany dplyr verbs respect grouping
Compare and contrast with the results we obtain if we omit rowwise(), where the mean column contains the averages of the three columns overall, rather than for each date and organisation:
# A tibble: 12,765 × 7
period org_code type attendances breaches admissions mean
<date> <fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 2017-03-01 RF4 1 21289 2879 5060 2450.
2 2017-03-01 RF4 2 813 22 0 2450.
3 2017-03-01 RF4 other 2850 6 0 2450.
4 2017-03-01 R1H 1 30210 5902 6943 2450.
5 2017-03-01 R1H 2 807 11 0 2450.
6 2017-03-01 R1H other 11352 136 0 2450.
7 2017-03-01 AD913 other 4381 2 0 2450.
8 2017-03-01 RYX other 19562 258 0 2450.
9 2017-03-01 RQM 1 17414 2030 3597 2450.
10 2017-03-01 RQM other 7817 86 0 2450.
# ℹ 12,755 more rowsc_across()
There’s also a c_across() function to select columns that looks really promising for rowwise() work, but bafflingly it is extremely slow here, taking 50x longer than the equivalent mutate(). This is a known issue - “particularly for long, narrow, data”. So this code is switched off and provided here for information only - although do feel free to try it out if you don’t mind a ten second wait.
Summary functions: nth() / first() / last()
Get the nth, first, or last values. Very useful inside a summarise() or similar when you want to be sure that you’re going to return a sensible result.
# A tibble: 4 × 2
speaker comment
<chr> <chr>
1 steve b
2 emma c
3 steve d
4 emma f Acknowledgments
I’m extremely grateful to Pauline Ward (PHS) for suggesting the dataset used for this session.